The core technology of this Repository of Glyco-Enzyme Expression Constructs is the generation of recombinant forms of the glycosylation enzymes in soluble forms when possible. Each of the glycosylation enzymes has a different strategy for its subcellular localication, including in many cases membrane anchoring sequences.
Summary of the glycosylation enzyme domain structures and topologies.
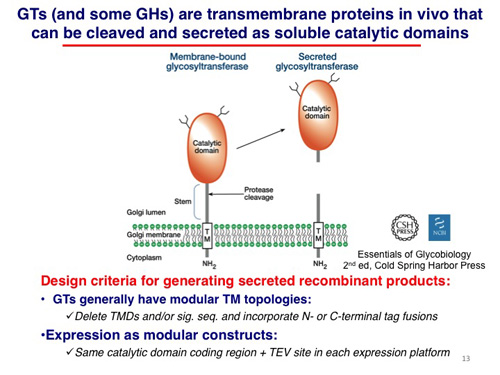
For each class of GT or GH sequence, truncation strategies were developed when possible to remove transmembrane segments to yield catalytic domains that could presumably be expressed in a soluble forms.
Summary of fusion protein strategies in mammalian and baculovirus vectors in the Repository.
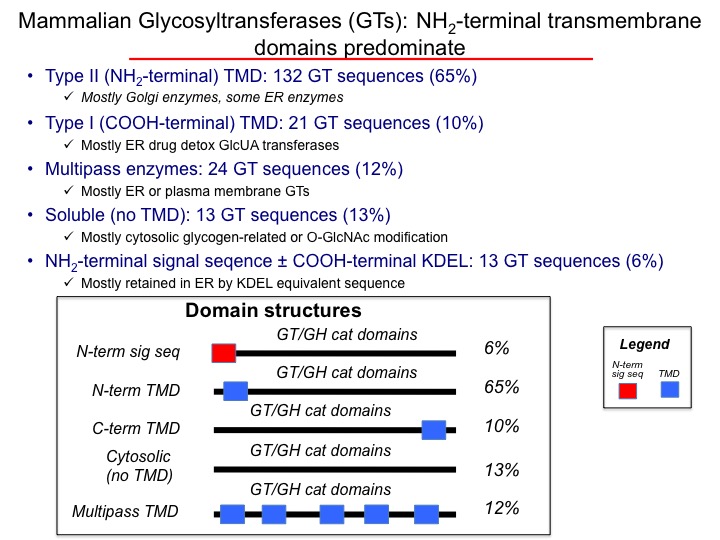
Mammalian glycosylation enzymes are generally found in membrane compartments of the secretory pathway. For glycosyltransferases (GTs) involved in the elaboration of glycan structures, the most common domain organization (~65%) is a catalytic domain facing the lumen of the endoplasmic reticulum or Golgi complex with a single transmembrane segment anchoring the protein to the membrane. In some instances proteolysis can occur in the luminal compartment to result in the release of a soluble catalytic domain that will progress through the secretory pathway and be secreted into the extracellular space.

Most GTs contain an NH2-terminal transmembrane anchor (65%) that acts as both a membrane insertion sequence for co-translational translocation into the membrane as well as acting as a membrane anchor (the proteins do not contain an additional N-terminal signal sequences). Some GTs (particularly the ER glucuronosyltransferases, ~10%of the total GTs) contain an NH2-terminal signal sequence and a COOH-terminal membrane anchor. A similar number of GTs are multipass transmembrane proteins (~12%), mostly in the ER. Finally, a number of GTs (~13%) do not appear to have a discernable transmembrane segment and reside in the cytosol.
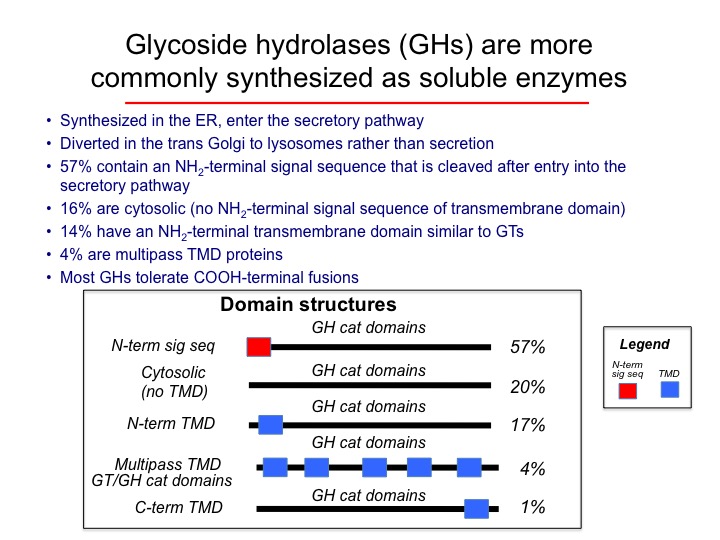
For glycoside hydrolases (GHs), the most common domain structure encodes an NH2-terminal signal sequence followed by a COOH-terminal catalytic domain (~57%). Most of these enzymes reside in lysosomes in mammalian cells. A smaller number of GHs contain NH2-terminal membrane anchor sequences without cleavable signal sequences (17%) and these proteins commonly reside in the Golgi complex. Finally, a number of GHs contain no discernable signal sequences or transmembrane segments and these proteins reside in the cytoso (20%)l.